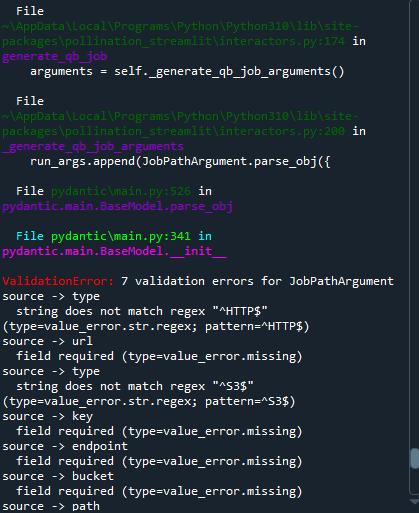
Thank you very much for the help. I have tried running simulations with the Python code and it works perfectly. But when I tried to input both the sim-par and the measures, I’m not quite understanding how it works. When I input the path of the sim-par (a sim-par that I had previously simulated in Pollination), it doesn’t work and gives me the following error:
@mostapha And focusing more on the measures part than the sim-par, what would be the best way to input custom measures for each model? Should I modify the OpenStudio Workflow (OSW) in Python with the exact value for each model?
Hi @batiste,
In both cases, it should be a path to the file or folder relative to the project folder on Pollination. That means that the file should be already available on Pollination before you pass it to the script. I suspect you are following the example here:
for files simply upload them and use the uploaded path.
sim_par_local_path = 'sim_par.json'
sim_par_path = new_study.upload_artifact(sim_par_local_path)
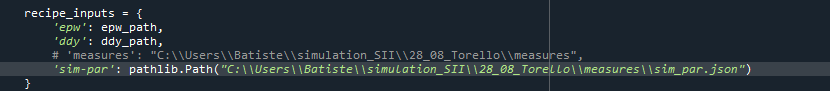
recipe_inputs = {
'sim=par': sim_par_path ,
}
For folders, you have to upload the files in the folder one by one. Then use the relative path to that folder as an input.
Hello @mostapha ,
Thank you very much for the clarification. I’m currently facing two issues when working with introducing measures, both in Grasshopper and through Python code.
In Grasshopper, the problem is that I’m not able to get each measure to change its output for each building in the GeoJSON. It’s as if when loading the measure, it’s given a list with input that gets modified for each measure. However, in the output, it only retains the value from the first input. It’s true that it generates as many output measures as the number of buildings (but all with the output value of the first building). Multiple studies measures inputs - #2 by Batiste - grasshopper - Ladybug Tools | Forum
Regarding Python, the issue is that I’ve tried setting up the ‘measures’ folder in Pollination using the get_all_file_paths and measure functions. There are two problems. First, the ‘measures’ folder isn’t set up in Pollination exactly as expected. It generates the main folder and subfolders correctly, but for the files inside, it creates both the file and a folder with the file’s name, and then duplicates the file within this folder. The second problem is related to the consequence of the Grasshopper issue. The measure values aren’t modified for the different buildings. Do I need to set up as many folders as there are buildings, each with the modified workflow.osw for each case? Or is there a way to modify only the workflow.osw file? I understand that since the folder creation and calculation don’t happen at the same time, the former option might be more suitable.
Here’s the code, which is just some quick modifications to read and set up the ‘measures’ folder. Then there are three functions to determine the inputs for the two measures and create the workflow.osw for each building.
import pathlib
import time
import requests
from requests.exceptions import HTTPError
import zipfile
import tempfile
import shutil
import urllib.parse
from typing import List
from pollination_streamlit.api.client import ApiClient
from pollination_streamlit.interactors import NewJob, Recipe, Job
from queenbee.job.job import JobStatusEnum
from honeybee_energy.measure import Measure
import json
import os
def tbd_input(any_constru):
any_constru = str(any_constru)
print(any_constru)
if any_constru == "40":
thermal_bridge_option = "spandrel (BETBG)"
elif any_constru == "60":
thermal_bridge_option = "poor (BETBG)"
elif any_constru == "80":
thermal_bridge_option = "uncompliant (Quebec)"
elif any_constru == "NBECT79":
thermal_bridge_option = "code (Quebec)"
elif any_constru == "CTE2006":
thermal_bridge_option = "regular (BETBG)"
elif any_constru == "CTE2013":
thermal_bridge_option = "spandrel HP (BETBG)"
elif any_constru == "CTE2019":
thermal_bridge_option = "efficient (BETBG)"
elif any_constru == "none":
thermal_bridge_option = "(non thermal bridging)"
else:
print(type(any_constru))
thermal_bridge_option = "uncompliant (Quebec)"
return thermal_bridge_option
def geojson_dict(geojson_path):
with open(geojson_path, 'r') as geojson_file:
geojson_data = json.load(geojson_file)
geojson_dict = {}
for feat in geojson_data['features']:
geojson_dict[str(feat['properties']['ids'])] = feat['properties']
return geojson_dict
def measure_clima(zona_climatica):
if zona_climatica:
folder_path = "C:\\Users\\Batiste\\Desktop\\ECLEKTE\\SUNO\\SUNO_SII\\SUNO_SII\\Data\\OutputData\\GroundTemp"
json_file = 'Ground_{}.json'.format(zona_climatica)
json_name = 'Ground_{}'.format(zona_climatica)
json_path = folder_path +"/"+ json_file
with open(json_path, 'r') as f:
ground_temp_set=json.load( f)[json_name]
measure_folder = "C:/Users/Batiste/Desktop/ECLEKTE/SUNO/SUNO_SII/SUNO_SII/Data/OutputData/measures"
measure_name_1 ="set_ground_temperatures_monthly"
measure_path = measure_folder + "/" + measure_name_1
measure = Measure(measure_path)
measure.arguments[0].value = ground_temp_set['temp_jan']
measure.arguments[1].value = ground_temp_set['temp_feb']
measure.arguments[2].value = ground_temp_set['temp_mar']
measure.arguments[3].value = ground_temp_set['temp_abr']
measure.arguments[4].value = ground_temp_set['temp_may']
measure.arguments[5].value = ground_temp_set['temp_jun']
measure.arguments[6].value = ground_temp_set['temp_jul']
measure.arguments[7].value = ground_temp_set['temp_aug']
measure.arguments[8].value = ground_temp_set['temp_sep']
measure.arguments[9].value = ground_temp_set['temp_oct']
measure.arguments[10].value = ground_temp_set['temp_nov']
measure.arguments[11].value = ground_temp_set['temp_dic']
else:
print('Error: No hi ha una zona climàtica definida')
return measure
def get_all_file_paths(directory):
file_paths = []
for root, _, files in os.walk(directory):
for file in files:
file_path = os.path.join(root, file)
file_paths.append(file_path)
return file_paths
def measure(measure_path, new_study):
all_files = get_all_file_paths(measure_path)
for file in all_files:
if 'workflow' not in file:
file = pathlib.Path(file)
relative_path = os.path.relpath(file, measure_path)
folder = os.path.dirname(relative_path).replace('\\','/')
folder_path = 'measure' + "/" + folder
print(file)
print(relative_path)
file_path = new_study.upload_artifact(file, target_folder= folder_path)
print(file_path)
def measures(geojson_dict, model_name,measures_path):
any_constru = geojson_dict[model_name]["id_clima"].split("_")[-1]
zona_climatica = "D1"
measure_ground = measure_clima(zona_climatica)
print('Ja tenim ground')
tbd_input_value = tbd_input(any_constru)
measure_folder = "C:/Users/Batiste/Desktop/ECLEKTE/SUNO/SUNO_SII/SUNO_SII/Data/OutputData/measures"
measure_name_2 = "tbd"
measure_path_2 = measure_folder + "/" + measure_name_2
measure_2 = Measure(measure_path_2)
measure_2.arguments[3].value = tbd_input_value
print('Ja tenim valor tbd')
measure_osw = measure_ground.to_osw_dict()
measure_osw_2 = measure_2.to_osw_dict()
osw = {
"steps": [
]
}
osw['steps'].append(measure_osw)
osw['steps'].append(measure_osw_2)
osw_path = measures_path.joinpath('workflow.osw')
with open(osw_path, 'w') as f:
json.dump(osw, f, indent=2)
print('Ja tenim osw actualitzat')
print(osw_path)
return osw_path
def submit_study(
study_name: str, api_client: ApiClient, owner: str, project: str, epw: pathlib.Path,
ddy: pathlib.Path, models_folder: pathlib.Path, geojson_dict: dict) -> Job:
print(f'Creating a new study: {study_name}')
# Assumption: the recipe has been already added to the project
recipe = Recipe('ladybug-tools', 'custom-energy-sim', '0.3.17', client=api_client)
input_folder = pathlib.Path(models_folder)
# create a new study
new_study = NewJob(owner, project, recipe, client=api_client)
new_study.name = study_name
new_study.description = f'Annual Energy Simulation {input_folder.name}'
# upload the weather files - you only need to upload them once, and you can use
# the path to them directly
assert epw.is_file(), f'{epw} is not a valid file path.'
assert ddy.is_file(), f'{ddy} is not a valid file path.'
epw_path = new_study.upload_artifact(epw, target_folder='weather-data')
ddy_path = new_study.upload_artifact(ddy, target_folder='weather-data')
sim_par = pathlib.Path("C:\\Users\\Batiste\\simulation_SII\\28_08_Torello\\sim_par.json")
sim_par_path = new_study.upload_artifact(sim_par)
measures_p = pathlib.Path("C:\\Users\\Batiste\\simulation_SII\\28_08_Torello\\measures")
measure(measures_p,new_study)
recipe_inputs = {
'epw': epw_path,
'ddy': ddy_path,
'sim-par': sim_par_path,
'measures': 'measure'
}
study_inputs = []
for model in input_folder.glob('*.json'):
model_name = model.stem
osw_dir = measures(geojson_dict, model_name,measures_p)
osw = pathlib.Path(osw_dir)
osw_path = new_study.upload_artifact(osw, target_folder='measure')
inputs = dict(recipe_inputs) # create a copy of the recipe
print(inputs)
# upload this model to the project
print(f'Uploading model: {model.name}')
uploaded_path = new_study.upload_artifact(model, target_folder=input_folder.name)
inputs['model'] = uploaded_path
inputs['model_id'] = model.stem # use model name as the ID.
study_inputs.append(inputs)
Hi @batiste,
This one is for @chriswmackey to help you with.
Can you share the link to a sample study on Pollination? I’m not sure if I understand the problem here. I should be able to help with this one.
I leave this one to @chriswmackey too.
Let’s wait for Chris and see what he thinks. It feels like with the amount of customization that you are trying to do and the scale of your studies we should work on the Python script. I’m not sure how measures need to be set up but once you get the folder structure correctly, everything should work as expected.
In addition, it would be great if you could share some sample files for testing. That will save me quite some time. Thanks!
This is one study implemented with this python script: Pollination Cloud App
I put an screenshot of one study:
And for the sample files:
28_08_Torello.zip (4.2 MB)
Thank you so much!
I sense there were some previous conversations here that I missed.
Are we still using the custom-energy-sim recipe to submit things to pollination? I cannot access the study that you linked to on the cloud app.
Assuming that you’re still using custom-energy-sim, are you making sure that you graft the inputs to the pollinate component? Remember that the typical workflow is to apply multiple measures to a single model. So just passing an un-grafted list of measures is going to apply them all to each model.
This is looking more and more like a case where we should just update the dragonfly-annual-energy-use recipe to accept mappers. That’s going to make the logic much simpler in terms of the order multiple measures get applied to each building and the values used as measure arguments for each building.
Thank you for your response @chriswmackey . The issue arises before the pollination component (custom-energy-sim). It comes from the Load Measure component. I’m not sure if it’s something related to the specific measure (TBD measure), but I’m providing a list with the values that the Measure needs to take for each building. However, in the output of the load measure, if I input 10 buildings, it gives me 10 measure outputs, but all with the values from the first input out of the ten. It’s as if it’s not updating the inputs. I have put some screenshots of the problema here Multiple studies measures inputs - #2 by Batiste - grasshopper - Ladybug Tools | Forum
Ok. Let’s discuss over on the LBT forum since this issue has more to do with LBT grasshopper mechanics than Pollination.
Regarding the issue of mappers with Dragonfly, you know @chriswmackey that you have a keen interest from me in this matter. This is just the beginning of the studies we want to conduct.
Furthermore, with the new features of DES, we are very interested in continuing the workflow towards the calculation of heat networks.
As for introducing measures from Python, I would like to know the correct methodology since my folders are getting duplicated. I understand that when it comes to applying different measures to different buildings, things get complicated, but the only way I can think of is creating separate ‘measures’ folders with the modified workflow.osw for each case.
If I manage to implement the differentiated measures from Python, it would be a significant step forward because I could quickly run the entire study instead of having to launch them in batches of 40 from Grasshopper.
Regarding sharing the study @mostapha , I’m not sure if it’s because it’s a company study, and I’m listed as a collaborator (plus, it’s private), so I’m not sure how I can send it to you.
Really, thank you very much to both of you. I’m learning a lot from this amazing platform, and I apologize for any persistence I may have shown. Thank you always for your time.